Welcome to PC-TraFF Server
News:
"PC-TraFF Extension" currently under review and will be provided here as soon as possible.
PC-TraFF: Potentially Collaborating Transcription Factor Finder
PC-TraFF_Professional is a complete web server that aims to identify potentially collaborating transcription factors (TFs) based on the locations of their binding sites in sequences.
Workflow of PC-TraFF_Professional:
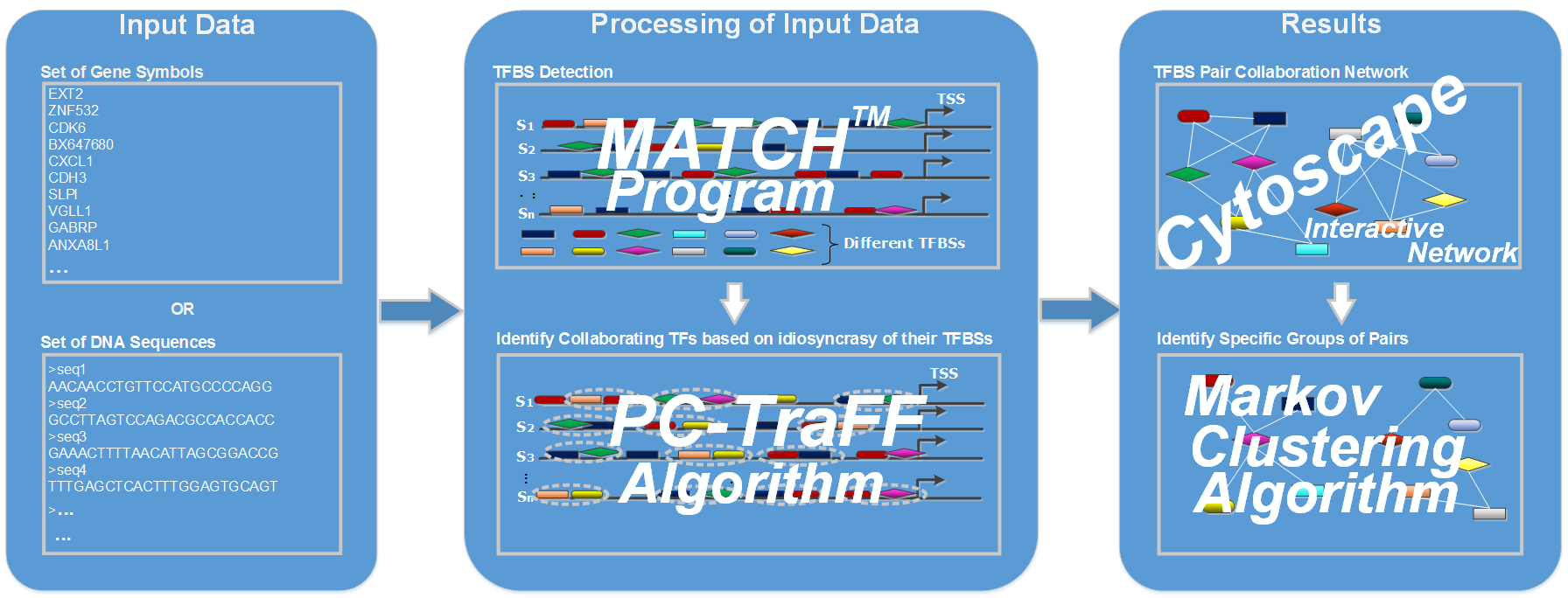
The binding of TFs frequently occurs in a cooperative manner due to their potential functional collaboration which leads to as cis-regulatory modules (CRMs). These modules are important for an effective regulation of the transcriptional machinery, even if they are not enriched in the corresponding promoter regions. The collaboration of TFs might stem from synergistic or antagonistic interactions between homotypic as well as heterotypic TFs. Such collaborations are likely to have effect on gene specificity and flexibility of the controlling of gene transcription ([PMID:18406148], [PMID:15930495], [PMID:22369666]).
References:
1. "PC-TraFF: identification of potentially collaborating transcription factors using pointwise mutual information," BMC Bioinformatics, vol. 16, no.1, pp. 1--21, December 2015.
2. "TFClass: a classification of human transcription factors and their rodent orthologs," Nucleic Acids Research, vol. 43, no.D1, pp.D97--D102, January 2015.
3. "The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation," Briefings in Bioinformatics, vol. 9, no.4, pp.326--332,, March 2008.
4. "MATCH: A tool for searching transcription factor binding sites in DNA sequences," Nucleic Acids Research, vol. 31, no.13, April 2003.